Aberrations in chromosome segregation during cell division lead to developmental defects and cancer. The centromere ensures faithful cell division in organisms. The centromere in eukaryotes is epigenetically marked by the presence of a specialized nucleosome wherein H3 is replaced by a variant called CENP- A. CenH3 containing nucleosomes are different from the canonical nucleosomes in structure, composition and function. Studying the specialized nucleosome may provide insights into the formation and regulation of the kinetochore on the centromere and hence give an important clue about the process of chromosome segregation. Other epigenetic regulation pathways due to the modifications occurring on the CenH3 tail may also govern the regulation of nucleosome and subsequently kinetochore formation.
In our lab, we aim to characterize the CenH3 proteins and reconstitute the CenH3 containing nucleosomes from budding yeast as well as humans to study the various structural, dynamics and functional aspects using solution and solid state NMR techniques along with biophysical methods. This will enable us to understand regulation of the specialized nucleosome formation and centromere function.
Team members working on the Specialized Nucleosome project are: Parveen Sehrawat, Rahul Shobhawat, Akashjyoti Pathak
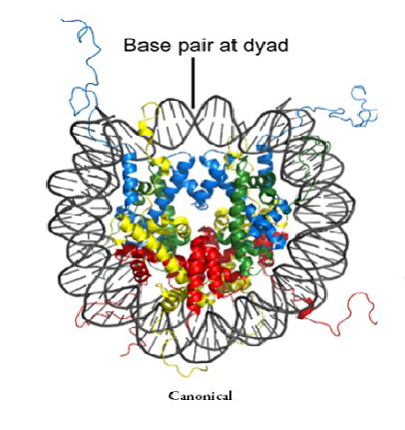
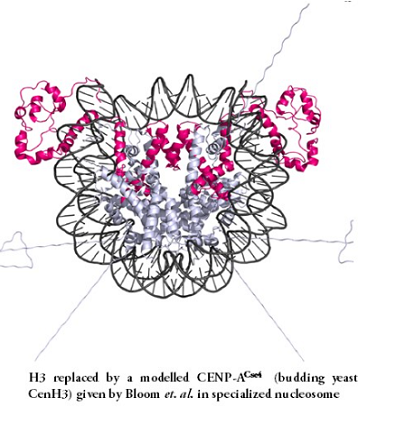
The Nucleosome Structure : Canonical Vs. Specialized
1. Implications of intrinsic disorder in specialized CenH3 variant CENP-ACse4 tail at the centromere
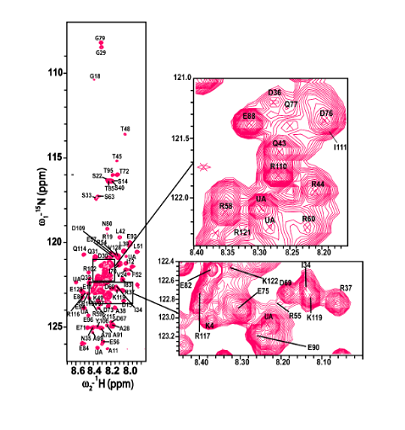
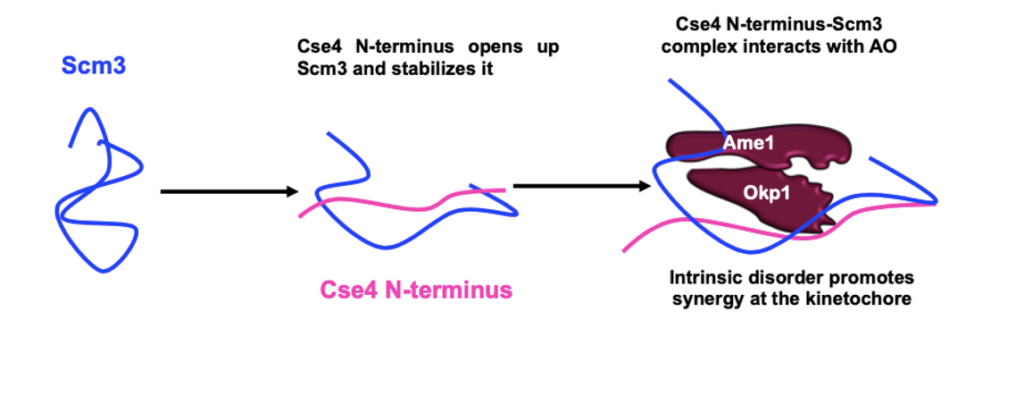
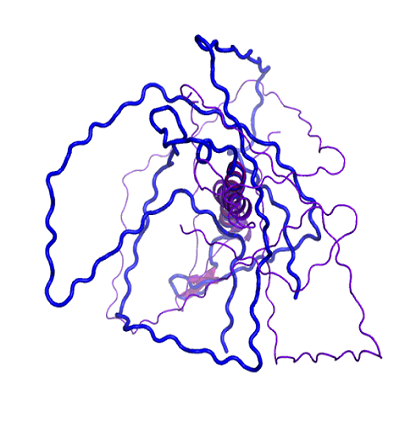
The kinetochore is a complex multiprotein network that assembles at a specialized DNA locus called the centromere to ensure faithful chromosome segregation. The centromere is epigenetically marked by a histone H3 variant – the CenH3. The budding yeast CenH3, called Cse4, consists of an unusually long N-terminal domain that has a role in kinetochore assembly. The N-terminus has not been characterized in all the available structural studies. We show that the N-terminus is intrinsically disordered.
The kinetochore has always been viewed as a largely ordered entity. However, secondary structure prediction studies show that most of the kinetochore proteins have considerable disorder in them. In our lab, we aim to unravel the implications of intrinsic disorder at the kinetochore with a focus on the CENP-ACse4 tail using solution-state NMR and biophysical tools.
Team members working on The Nucleosome Group project are: Shivangi Shukla and Anusri Bhattacharya

2. Implications of heavy metal compounds on nucleosome dynamics

Heavy metals interact with the H2A-H2B acidic patch to bring about nucleosome compaction which has been shown to cause apoptosis in cancer cells.
We aim to decipher the mechanism of palladium-based compounds on the nucleosome dynamics to provide rationale for a drug design which can be used as a potential treatment in cancer patients.
Team members working on This project are: Parveen and Rahul Shobhawat