Advances in Proteomics Technologies
(APT -2024)
Workshops and International Conference
February 17-20, 2024
IIT Bombay, Mumbai


Prof. Sanjeeva Srivastava
IIT Bombay
Convener’s Message
The field of proteomics aims to explore proteins, delving into their structure, function, and interactions within biological systems. Comprehending the proteome stands as a crucial factor in grasping cellular function, disease pathology, and the development of targeted therapies. Proteomics offers valuable insights into the molecular mechanisms that underlie various physiological and pathological processes, aiding in the identification of potential drug targets. Recent strides in Mass Spectrometry have enabled the identification of key proteins governing cellular processes, their signaling pathways, and abnormalities leading to disease development.
The rapid evolution of instrumentation technology has significantly propelled the field of proteomics. Approaches in proteomics have seen improvements across various levels, including sample preparation, MS data acquisition, and data analysis. Emerging fields such as cell surface proteomics, spatial proteomics, and single-cell proteomics, though in their early stages, show considerable promise and are poised to play a pivotal role in advancing our understanding of biology in the years to come.
In addition to mass spectrometry-based approaches, alternative methods like protein sequencing and array-based approaches are yielding promising results. Techniques such as surface plasmon resonance and Bio-layer interferometry are increasingly utilized in Biopharma, where they play a pivotal role in identifying and characterizing biosimilars.
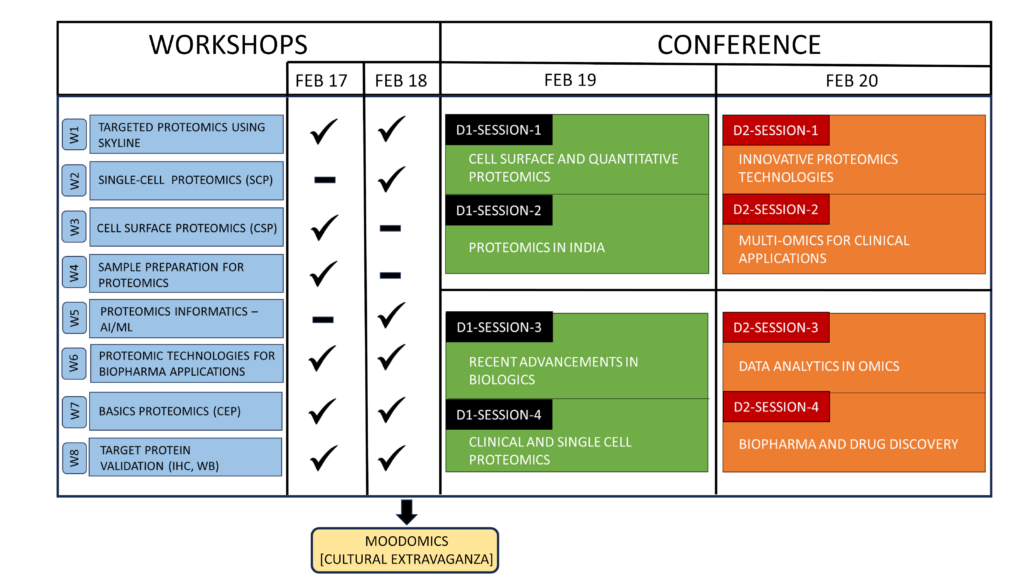
Two-day workshops (7 parallel tracks, Feb 17-18) and two-day conference (Feb 19-20) aim to provide participants with insights into the recent advances in the field of proteomics, promising and exciting decade ahead.
Recent News & Updates
APT-2024 ROADMAP
EVENT FLYERS
DISTINGUISHED SPEAKERS
Alexander Makarov
Thermo Fisher Scientific,
Germany
Albert Heck
Utrecht University, Netherlands (Online)
Arun Wiita
UCSF, USA
Bernd Wollscheid
ETH Zurich, Switzerland
Bernhard Kuster
Technical University Munich





Brendan MacLean
University of Washington, USA
Christina Ludwig
TUM, Germany
Fuchu He
Beijing Proteome Research Center, China (Online)
Graham Roy Ball
Anglia Ruskin University, UK
Hanno Steen
Harvard Medical School, USA





Jim Palmeri
ProtiFi
Jim Wells
UCSF, USA (Online)
Jochen Schwenk
Scilife Lab,
Sweden (Online)
John Wilson
ProtiFi, USA
Jonathan Blackburn
University of Cape Town, SA (Online)





Judith Steen
Harvard Medical School, USA
Mahesh Bhalgat
Veeda Clinical Research
Matthias Mann
Max Planck Institute of Biochemistry, Germany
Neil Kelleher
Northwestern University, USA (Online)
Nikolai Slavov
Northeastern University, USA (Online)





Parag Mallick
Stanford University,
USA (Online)
Prashant Kumar
Karkinos Healthcare Pvt. Ltd
Rajiv Sarin
ACTREC, India
Ravi Kannan
CCHRC, India
Robert Moritz
Institute for Systems Biology, USA





Rosa Viner
Thermo Fisher Scientific,
USA (Online)
Ruth Huttenhain Stanford University, USA (Online)
Shourjo Ghose
Bruker Daltonics
Tiannan Guo
Westlake University, China (Online)
Thierry Nordmann
Max Planck Institute of Biochemistry, Germany (Online)
PROTEOMICS FOR BIOPHARMA





Ashutosh Kumar
IIT Bombay
Bhupesh Dewan
Zuventus Healthcare Ltd.
Dinesh Palanivelu
ThermoFisher Scientific
Kamal Mandal
Gujarat Biotechnology University
Narendra Chirmule
SymphonyTech Biologics





Ratnesh Jain
Institute of Chemical Technology
Ravi Shankara
Sun Pharma
Ravi Trivedi
Zydus
Saravanan Kumar
ThermoFisher
Scientific
Ravi Krovidi
Ingenuity Biosciences Pvt. Ltd.
PROTEOMICS IN INDIA





Amol Suryavanshi
Institute of Life Sciences
Bhubaneshwar
Geetanjali Sachdeva
NIRRH Mumbai
Mahesh Kulkarni
NCL Pune
Niranjan Chakraborty
NIPGR
New Delhi
Prasanna Venkatraman
ACTREC Mumbai





Ramesh Ummanni
CSIR IIST Hyderabad
Ravi Sirdeshmukh
Institute of Bioinformatics Bangalore
Shantanu Sengupta
IGIB Delhi
Srikanth Rapole
NCCS Pune
Subhra Chakraborty
NIPGR
New Delhi



Suman Thakur
CSIR CCMB Hyderabad
Surekha Zingde
IWSA
T S Keshava Prasad
Yenepoya University Mangalore
EARLY CAREER RESEARCHERS (ECR)





Ankit Jain
NCBS
Arup Acharjee
University of Allahabad
Jubilee Purkayastha
DRDO
Kartiki Desai
NIBMG
Mukunda Goswami
Central Institute of Fisheries Education





Pratibha Sharma
IHBAS Delhi
Rashmi Rana
Sir Ganga Ram Hospital
Sandipan Ray
IIT Hyderabad
Sumit K. Singh
IIT BHU
Surya Pratap Singh
IIT Dharwad

Syed K. Hasan
ACTREC Mumbai
WORKSHOP SPEAKERS





Aditi Jain
ACS
Balaji Sitharaman
Millennial Scientific, USA
Harsharan Singh Bhatia
Helmholtz Munich, Germany
Mayuri Gandhi
IIT Bombay
Moumita Basu
Thermo Fisher Scientific

Shrivalli R Kamath
Thermo Fisher Scientific
WORKSHOPS
Quantitative Mass Spectrometry based proteomics approaches have been undergoing huge improvements over the last decade owing to advances in instrumentation and sample preparation methods. These have been largely driven by shotgun approaches and targeted approaches. The shotgun approaches are also known as data dependent discovery proteomics approaches, and they have been used to quantify proteomes from various sample types. The targeted approaches such as Selected Reaction Monitoring (SRM) and Parallel Reaction Monitoring (PRM) are the method of choice for quantifying proteins of low abundance with a high degree of accuracy in clinical isolates as well as other complex sample types. These approaches have also overcome the limited scalability and reproducibility of shotgun approaches but suffer from identification of only a limited number of proteins per run making them more suitable for Hypothesis-based research or validation studies. The last few years have seen the emergence of a new approach called the Data Independent Acquisition (DIA). In this approach, the fragment ion spectra are systematically acquired using deterministic peptide ion isolation windows spanning the entire mass range expected to house most digested peptides. Latest developments in ion-mobility, new acquisition schemes, data analysis workflows and software tools have propelled the use of DIA towards consistent quantification of thousands of proteins from complex sample cohorts. The targeted proteomics workshop aims to introduce to the participants the quantitative approaches currently driving proteomics-based research and data analysis using Skyline. Participants are expected to have a basic to moderate understanding of Skyline. The workshop will also briefly touch upon the use of plug-ins for Skyline and statistical analysis using MS Stats. The instructors will also speak about the aspects involved in method development and troubleshooting. Lastly, the participants will be encouraged to plan their own experiments and discuss them with the experts.

Brendan MacLean
Mr. Brendan MacLean is a Senior Software Engineer in the laboratory of Mike MacCoss at the University of Washington, Department of Genome Sciences. He has been responsible for all aspects of design, development and support in creating the Skyline Targeted Mass Spec Environment with an ever-increasing user base around the world.

Christina Ludwig
Dr. Christina Ludwig is the Head of the Proteomics Unit at the Bavarian Center for Biomolecular Mass Spectrometry (BayBioMS), Technical University Munich. Prior to joining BayBioMS she worked as a senior scientist at the lab of Prof. Ruedi Aebersold at ETH Zurich. Here she was instrumental in developing advanced proteomics approaches like Multiple reaction monitoring and SWATH-MS. She also played a key role in designing the targeted proteomics course originally started at ETH-Zurich and is among the few instructors who have continued to play a key role in training researchers in targeted proteomics for over a decade now.
Complex biological interactions are a result of the dynamic interplay between different cell types found inside the body. The differences in functional and morphological features of the different cell types are also dependent on the complex interplay between the genome, transcriptome, proteome, and other regulatory molecules that make up the cell. Single cell genomics and transcriptomics have long been at the forefront of single cell omics research allowing researchers to study the mutations at the cellular level. However, the effects of these mutations can only be understood if there is information about the proteome level changes. Single cell proteomics is the latest branch of omics approaches aimed at understanding alterations at the single cell resolution. Despite initial challenges of analysing only small number of proteins, with advanced mass spectrometry technologies, community is now aiming to achieve ~ 5000 proteins from a single cell, a number which is comparable to global proteomics data. Since the approaches do not employ an amplification step unlike the other approaches, efforts are on to develop more effective and efficient extraction protocols to minimize protein loss during sample processing. Additionally, these approaches suffer from lack of consensus for experimental design, data analysis, and interpretation. However, despite these challenges, single cell proteomics approaches hold immense potential and are already helping researchers understand cellular heterogeneity in complex tissues and tumors. The single cell proteomics workshop aims at introducing participants to the important sample preparation approaches and data analysis pipelines commonly used by researchers around the world. The instructors will provide an insight into the application of single cell proteomics for understanding spatial and subcellular heterogeneity in complex tissue types. Due to the highly specialized nature of the field, participants with moderate to advanced knowledge in proteomics will be given preference for participation in the workshop.

Hanno Steen
Dr. Hanno Steen is an Associate Professor of Pathology at Harvard Medical School and Principal Investigator at Boston Children’s Hospital. His lab works on novel methods for identifying and quantifying various protein modifications that are involved in pediatric diseases in order to provide new insights into the underlying biological processes.

Judith Steen
Dr. Judith Steen is an Associate Professor of Neurobiology at Harvard Medical School, a member of the Harvard Stem Cell Institute and the Director of the Neuroproteomics Laboratory in the F. M. Kirby Neuroscience Center at Boston Children’s Hospital. Her laboratory works on understanding neuroregeneration and neurodegenerative diseases using systems biology approaches.

Shourjo Ghose
Shourjo Ghose received his Ph.D from Montana State University in Biochemistry. He proceeded to do his post doctoral studies at the Scripps research institute . Shourjo joined Bruker Daltonics in 2017 as an appalications scientist specializing in performing proteomics measurements on the timsTOF line of systems. In 2020, Shourjo was promoted to be a an account manager , responsible for the New England region in North America. As of 2022, Shourjo is the Proteomics Business Unit manager responsible for the North American business.

Proteins are dynamic biomolecules involved in a wide variety of functions in biological systems ranging from catalytic reactions, overall development and maintenance of cells, cell to cell communication, etc. to name a few. However, despite their wide-spread occurrence in the body, only 30% of the total proteins can be found on the cell surface and at levels much lower than the proteins found inside the cell. Incidentally, the proteins found on the cell surface make up to approximately 65% of the FDA-approved therapeutic targets, highlighting the need to better understand and characterise these proteins. The study of the alterations found in the cell surface proteins commonly referred to as ‘Surfaceome’ has widespread implication in the field of immunobiology. Studies aimed at providing a holistic understanding of the alterations of these proteins typically deal with investigating biological signatures driven by protein post-translational modifications, protein-protein interactions, and altered sub-cellular localization. These have provided a fresh outlook towards complex diseases like blood cancers resulting in development of new-age therapeutic approaches. The knowledge from these studies if coupled with synthetic immunology tools to make targeted therapeutics, can pave way for precise and targeted treatments in patients. However, such proteomics approaches are challenging due to the relatively low abundance of the cell surface proteins and often require specialised sample preparation strategies and novel data analysis pipelines. The Cell surface proteomics workshop has been specifically designed to address the rapidly growing demand for better understanding of immune cells in complex diseases like cancers and other neurodegenerative disorders. The workshop will introduce participants to this new field by combining concepts of chemical biology with mass spectrometry-based proteomics. It will also familiarise participants the pipeline, going all the way from target discovery to therapeutic development to clinical trials. Due to the interdisciplinary nature of the field, participants are expected to have a basic understanding of proteomics, bioinformatics, chemical biology, and mechanistic biology.

Arun Wiita
Dr. Arun Wiita is a Clinical Pathologist and physician-scientist at the Department of Laboratory Medicine, University of California San Francisco. His lab is majorly involved in the use of targeted and quantitative proteomics approaches to answer important biological questions in myeloma and B-cell leukemia.

Bernd Wollscheid
Dr. Bernd Wollscheid is a Professor at Department of Health Sciences and Technology, ETH Zurich. The goal of his laboratory is to functionally understand the cellular surfaceome and its signaling islands as a complex information gateway connecting the intracellular to the extracellular interactome.

Rosa Viner (Online)
Dr. Rosa Viner holds a Ph.D. in Biochemistry and Biophysics from the Moscow State University, Russia and is currently the Senior Vertical Manager for Proteomics at Thermo Fisher Scientific in San Jose, CA. She joined Thermo Fisher Scientific in 2004 and is involved in all areas of proteomics research with specific focus on relative quantification, PTM characterization and integrated structural biology workflows. She serves as liaison between Life Science mass spectrometry and Pierce Protein research business units to provide complete solutions/workflows to customers.

Sample preparation is among the most crucial steps for the success of an omics experiment and accounts for the largest source of data variability across experiments. Various sample processing strategies revolve around the same basic steps starting with the careful lysis of the cells to recover the biomolecules of interest, followed by enrichment of the specific biomolecules and their downstream processing. For proteomics sample preparation, the steps include physical or chemical lysis of the cells in a lysis buffer, successful recovery of proteins either by separating the cell debris or by precipitation, followed by reduction, alkylation and digestion of proteins, and clean-up or desalting of the digested peptides prior to LC-MS/MS analysis. Sample preparation methods differ in the composition of lysis buffers, the strategies used for cell disruption and lysis, denaturing conditions used during digestion and methods used for sample clean up. While it is assumed that the methods used for cell disruption and lysis are universal, sample processing approaches for proteomics experiments require the use of buffers and reagents that are compatible with the LC-MS/MS analysis. Hence, it is essential to optimize and choose the right sample preparation method depending on the sample of interest, availability of reagents, cost-effectiveness, and difficulty of the protocol. The last few years have seen a rise in specialized sample preparation approaches which are aimed at reducing the steps involved in the workflow and are highly reproducible in their output. One such approach is the S-trap sample processing technology that allows the use of SDS in the lysis buffer thereby resulting in the recovery of even poorly soluble proteins like membrane proteins. Another approach that has been gaining popularity for its relative ease of usage and short processing time is the preomics improved sample technology (iST). Using proprietary reagents, this method allows sample preparation within two hours through a manual, semi or fully automated approach. This method also allows processing of multiple samples at once and requires very low amounts of starting material which is a major advantage while working with clinical samples. The sample preparation workshop has been designed specifically for researchers in the early stages of their proteomics journey with emphasis on choosing the most effective sample preparation strategy for maximal identification of proteins. The participants of this workshop will also be introduced to the most observed problems during sample preparation and possible ways to troubleshoot the problems. Additionally, the participants will be educated about the commercially available sample preparation kits for faster and more efficient sample preparation.

John Wilson
Dr. John Wilson is a CEO, an entrepreneur, a PhD scientist, and an expert in the fields of protein and biomolecule analysis, biomarkers and mass spectrometry. He is founder of Protifi, a spinout from the Cold Spring Harbor Laboratory, which makes robust and reliable sample preparation systems for a wide range of sample types. His company also helps take R&D from the bench to the clinic, helps determine states of health or disease, or predict drug responses. Besides Protifi, Dr. Wilson has also founded NYC Bio and can be seen wearing many hats! In addition to all this, he takes pride in passing the knowledge he has acquired during his time at premier institutes like Rockefeller university, Stanford university, Universität Tübingen and the Cold Spring Harbor Laboratory to young students and researchers.
The last two decades have seen an unprecedented growth in the use of mass spectrometry (MS)- based methods for exhaustive identification and quantification of proteins in biological samples. The rapid advancements in instrumentation, coupled with improvements in sample preparation strategies have led to increased coverage of proteins. This has in-turn made data analysis workflows more complex necessitating the use of greater computational power for handling the resulting datasets. The proteomics community has been blessed with the availability of numerous softwares that can handle large and complex datasets. While most of the softwares are good in handling data from shotgun experiments acquired using data dependant acquisition (DDA) approaches, only a handful of softwares are efficient in handling complex data arising from use of data independent approaches (DIA). To make matters worse, most of these software capable of handling DIA data are not freely available and require a license. Data analysis from proteomics experiments also requires pre-processing prior to statistical testing, network analysis and data visualization. As such there is no one method for these steps and the researchers are expected to have a basic understanding of the various approaches most suited for proteomics data analysis. The emergence of Machine Learning (ML) and Artificial Intelligence (AI) has also accelerated development of proteomics data analysis softwares and tools commonly used to data analysis. The workshop on Omics data analysis will provide participants with a basic understanding on the problems commonly encountered with proteomics datasets and the strategies one must employ while analysing complex datasets. The workshop will also briefly touch upon the role of ML and AI in data processing providing participants an idea of the potential of these approaches in handling large datasets. Lastly, the participants will be introduced to a few commonly used databases, including BrainProt, and tools for network analysis and data visualization.

Graham Roy Ball
Dr. Graham Ball is a Professor at Anglia Ruskin University and a visiting Professor at the Indian Institute of Technology Bombay. His current research interests are focused on the development and application of machine learning algorithms using Artificial Neural Networks (ANNs) to medical diagnostics, systems biology, and drug target discovery.

Tiannan Guo (Online)
Dr. Tiannan Guo is a Professor at the School of Life Sciences at Westlake University where he leads a multi-disciplinary team to develop reproducible, fast, deep, and low-cost proteomics technologies. His lab focuses on developing computational resources to democratize the data analysis of proteomic big data sets by using a combination of pressure-cycling technology and data-independent acquisition. The lab also develops cutting edge proteomics technologies to precisely quantify maximum number of proteins from the minimum amount of biological or clinical samples with the maximum sample throughput, enabling the generation of proteomic big data research to tackle complex biomedical questions.
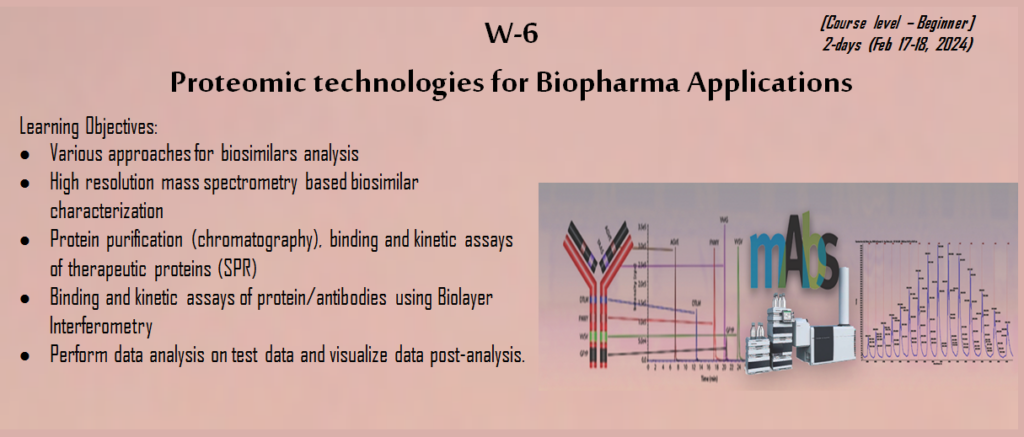
Since the last COVID-19 pandemic India have seen an increasing demand for biopharmaceuticals and biosimilar due to an increasing prevalence of chronic and febrile diseases, aging population with multiple comorbidities and ultimately shifting of paradigm into better healthcare, point of care and focused quality of life. Both Academia and Industry led research collaboration have led to development of novel biopharmaceutical products, which made India key player in global biosimilars market in field of therapeutics such as oncology, rheumatology, and diabetes. The current workshop and conference at APT-2024 will help you in understanding the latest cutting-edge applications biosimilars and revolutionize in-depth insights into the characterization, formulation and manufacturing Pipeline for Biopharma market in India. In the workshop you will be trained in Mass Spectrometry based identification of metabolites and small molecules, their targeted validation, dilution linearity, and other quantification aspect with Ligand binding Assay using SPR/ BLI which are widely used in Pharma and Biopharma Industry. In the conference you will provided insights to mAbS Engineering, Biosimilar characterization, Cell and gene therapy, and Big data, AI and ML based application in Drug discovery and Biotherapeutics. This will be a great opportunity for (Researchers and Scientists, Academia and Students, Pharmaceutical Industry Enthusiasts and Regulatory Affairs Professionals) to be connected with like-minded professionals, researchers and industry experts, which will further result in valuable collaborations, exchange ideas, and expand your professional network within the vibrant biosimilars community. Join us at the Biosimilars Workshop APT 2024, where unison of innovation and knowledge leads to the future of biopharmaceuticals

Arghya Banerjee
Dr. Arghya’s research focuses on studying the pathobiology of Pituitary Adenomas using the Proteo-Metabolomics approach. He has also worked in SARS-CoV-2 severity prediction using various spectrometry approaches
More speaker details are coming soon…
The last decade has seen an increase in the interest towards specialized proteomics applications. Sample preparation plays an important role in the success of a proteomics experiment. The basic proteomics workshop will familiarize participants with the recent advancements in sample preparation with emphasis on the nuances and steps requiring attention, introduce the recent advancements in method development and data acquisition approaches, and finally acquaint with data pre-processing and analysis approaches. The strategies and approaches for preparing a clean sample evolve at a rapid pace owing to advancements in the field. Quantitative proteomics approaches have seen a rapid growth in recent times, thanks to the developments in MS instrumentation. The developments in sample preparation strategies, liquid chromatography and mass spectrometry instrumentation have together contributed to increased reliability, coverage, and reproducibility of proteomics approaches. Following data acquisition, a typical mass spectrometry experiment involves analysis of the acquired datasets. While most commercial softwares for MS data analysis are expensive, there are many freely available softwares for use. A thorough understanding of the data complexity and the commonly associated problems with mass spectrometry data is necessary for effective data analysis. The workshop has also been designed to cater to the ever-growing need for development of skillset and knowledgebase in proteomics approaches, with emphasis on hands-on training from field-leading experts. The workshops will provide participants the opportunity to interact with experts during the course besides visiting our state-of-the-art proteomics facility.
The last decade has seen an increase in the interest towards specialized proteomics applications. Sample preparation plays an important role in the success of a proteomics experiment. The basic proteomics workshop will familiarize participants with the recent advancements in sample preparation with emphasis on the nuances and steps requiring attention, introduce the recent advancements in method development and data acquisition approaches, and finally acquaint with data pre-processing and analysis approaches. The strategies and approaches for preparing a clean sample evolve at a rapid pace owing to advancements in the field. Quantitative proteomics approaches have seen a rapid growth in recent times, thanks to the developments in MS instrumentation. The developments in sample preparation strategies, liquid chromatography and mass spectrometry instrumentation have together contributed to increased reliability, coverage, and reproducibility of proteomics approaches. Following data acquisition, a typical mass spectrometry experiment involves analysis of the acquired datasets. While most commercial softwares for MS data analysis are expensive, there are many freely available softwares for use. A thorough understanding of the data complexity and the commonly associated problems with mass spectrometry data is necessary for effective data analysis. The workshop has also been designed to cater to the ever-growing need for development of skillset and knowledgebase in proteomics approaches, with emphasis on hands-on training from field-leading experts. The workshops will provide participants the opportunity to interact with experts during the course besides visiting our state-of-the-art proteomics facility.

John Wilson
Dr. John Wilson is a CEO, an entrepreneur, a PhD scientist, and an expert in the fields of protein and biomolecule analysis, biomarkers and mass spectrometry. He is founder of Protifi, a spinout from the Cold Spring Harbor Laboratory, which makes robust and reliable sample preparation systems for a wide range of sample types. His company also helps take R&D from the bench to the clinic, helps determine states of health or disease, or predict drug responses. Besides Protifi, Dr. Wilson has also founded NYC Bio and can be seen wearing many hats! In addition to all this, he takes pride in passing the knowledge he has acquired during his time at premier institutes like Rockefeller university, Stanford university, Universität Tübingen and the Cold Spring Harbor Laboratory to young students and researchers.

Sanjeeva Srivastava
Dr. Sanjeeva Srivastava, Ph.D., is the head of the proteomics facility and a professor at the Indian Institute of Technology Bombay. Currently, he is appointed as a full-time visiting professor at the Department of Laboratory Medicine, School of Medicine at the University of California, San Francisco. His research on protein biomarkers of infectious diseases and brain tumors has resulted in 150+ publications & over 20 patents filed, including Nature and Cell press publications.
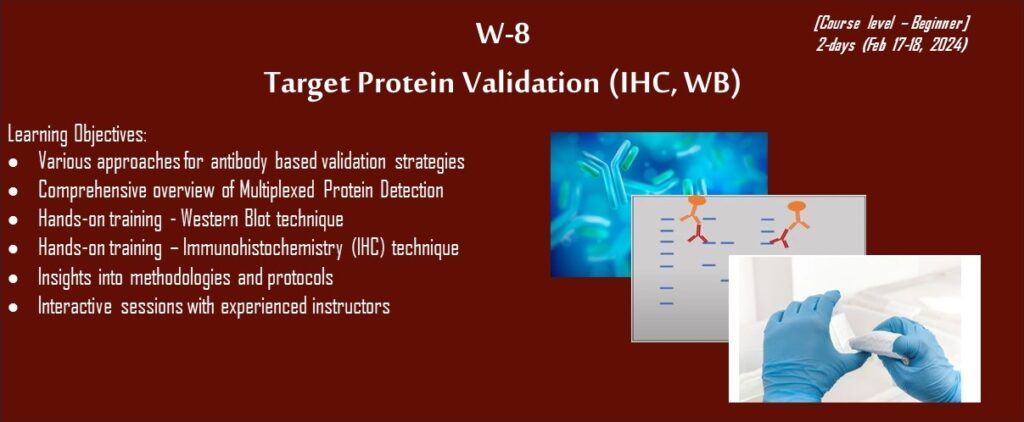
Unlocking the nuances of antibody-based validation strategies, mastering Immunohistochemistry (IHC), and Western Blot techniques is paramount for researchers endeavoring to translate targets from proteomics experiments. These methods not only present a visual portrayal of protein expression in tissues (IHC) but also furnish a means for quantitative analysis of protein levels (Western Blot). Proficiency in these techniques ensures the precision and dependability of protein data, thereby fortifying the validity of research findings. Our workshop empowers participants with indispensable skills to navigate the intricacies of protein validation, elevating the robustness and credibility of their research outcomes in the pursuit of translational excellence.

Moumita Basu
Moumita Basu is a R&D Scientist in Protein Biology at Thermo Fisher Scientific. After obtaining her BS-MS degree in Biological Sciences from the Indian Institute of Science Education and Research, Bhopal, she received her Ph.D. in Molecular Biology from Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore in 2022. Her research focused on understanding the crosstalk between transcription and epigenetic machinery and implication of their dysregulation in manifesting various pathophysiological conditions such as stress, carcinogenesis, etc. in cellular and animal models as well as patient samples. Since joining Thermo Fisher Scientific in 2022, she has focused on immuno-application work to support the development and marketing of new technologies.

Shrivalli R Kamath
Shrivalli R Kamath is a R&D Scientist in Protein Biology at Thermo Fisher Scientific. She completed her bachelors degree in Engineering in Biotechnology from NMAMIT, Nitte, Karnataka and her post-graduate diploma in Cellular and Molecular Diagnostics from School of Life Sciences, Manipal, Karnataka. Since joining Thermo Fisher Scientific in 2015, she has focused on the functional validation of primary and secondary antibodies in western blotting, immunocytochemistry and immunohistochemistry applications.
CONFERENCE

The rapid advancements in instrumentation technology have provided a major boost to the field of proteomics. Proteomics approaches have also undergone changes and improvements at levels ranging from sample preparation, MS data acquisition and data analysis. Emerging fields like cell surface proteomics and single cell proteomics, though in their nascent stages, have already shown a lot of promise and will play a key role in advancing our understanding of biology in the years to come. In addition to mass spectrometry-based approaches, alternative approaches such as protein sequencing have also been gaining speed and hold promise for application in other associated fields. Approaches like surface plasmon resonance and Bio-layer interferometry are increasingly finding use in Biopharma where their role has been pivotal in identification and characterization of biosimilars. This 2-day conference (Feb 19-20, 2024) having two simultaneous tracks – “Proteomics for Biologists” and “Proteomics tools for Biopharma” intends to provide participants knowledge of the recent advances in field of proteomics with a promise of an exciting decade ahead.
Conference Schedule (Feb 19-20, 2024)
(Nearly final — updated Jan 31, 2024)
Day 1-19th February, 2024
| 8:20 AM | Prof. Sanjeeva Srivastava | Indian Institute of Technology Bombay | Welcome |
| 8:25 AM | Prof. Fuchu He | Beijing Proteome Research Center, China | Plenary (Online) |
| 8:50 AM | Prof. Jim Wells | University of California San Francisco, USA | Plenary (Online) |
| 9:15 AM | Prof. Arun Wiita | University of California San Francisco, USA | Plenary |
| 9:43 AM | Prof. Bernd Wollscheid | ETH Zurich, Switzerland | Plenary |
| 10:11 AM | Prof. Judith Steen | Harvard Medical School, USA | Plenary |
| 10:39 AM | Dr. Christina Ludwig | Technical University of Munich, Germany | Plenary |
| TEA | |||
| 11:25 AM | Dr. Prasanna Venkatraman | Advanced Centre for Treatment, Research and Education in Cancer (ACTREC), Mumbai | Invited |
| 11:40 AM | Prof. Ramesh Ummanni | Indian Institute of Chemical Technology (CSIR-IICT) Hyderabad | Invited |
| 11:55 AM | Dr. Amol Suryavanshi | Institute of Life Sciences Bhuvaneshwar | Invited |
| 12:10 PM | Prof. Shantanu Sengupta | Institute of Genomics and Integrative Biology, Delhi | Invited |
| 12:25 PM | Prof. Srikant Rapole | National Centre For Cell Science, Pune | Invited |
| 12:40 PM | Prof. Subhra Chakraborty | National Institute of Plant Genome Research (NIPGR), New Delhi | Invited |
| 12:55 PM | Dr. Suman Thakur | Centre for Cellular and Molecular Biology (CCMB), Hyderabad | Invited |
| 1:10 PM | Dr. Rashmi Rana | Sir Ganga Ram Hospital Delhi | ECR |
| LUNCH | |||
| 2:05 PM | Prof. Ravi Sirdeshmukh | Institute of Bioinformatics Bangalore | Invited |
| 2:20 PM | Dr. T S Keshava Prasad | Yenepoya University Mangalore | Invited |
| 2:35 PM | Prof. Niranjan Chakraborty | National Institute of Plant Genome Research (NIPGR), New Delhi | Invited |
| 2:50 PM | Dr. Narendra Chirmule | SymphonyTech Biologics | Workshop |
| 3:20 PM | Dr. Dinesh Palanivelu | ThermoFisher Scientific | Invited |
| 3:35 PM | Dr. Ravi Trivedi | Zydus Lifesciences | Invited |
| 3:50 PM | Prof. Albert Heck | Utrecht University, Netherlands | Plenary (Online) |
| 4:15 PM | Prof. Surya Pratap Singh | Indian Institute of Technology Dharwad | ECR |
| 4:40 PM | Prof. Syed K. Hasan | Advanced Centre for Treatment, Research and Education in Cancer (ACTREC), Mumbai | ECR |
| 4:50 PM | Prof. Jonathan Blackburn | University of Cape Town, South Africa | Plenary (Online) |
| 5:15 PM | Prof. Bernhard Kuster | Technical University of Munich, Germany | Plenary (Online) |
| 5:40 PM | Prof. Nikolai Slavov | Northeastern University, USA | Plenary (Online) |
| 8:00 PM | Quiz-Online (for the participants) |
Day 2-20th February, 2024
| 8:25 AM | Prof. Sanjeeva Srivastava | Indian Institute of Technology Bombay | Welcome |
| 8:30 AM | Prof. Ruth Huttenhain | Stanford University, USA | Keynote (Online) |
| 8:50 AM | Prof. Neil Kelleher | Northwestern University, USA | Plenary (Online) |
| 9:15 AM | Prof. Parag Mallick | Stanford University, USA | Plenary (Online) |
| 9:40 AM | Prof. Matthias Mann | Max Planck Institute of Biochemistry, Germany | Plenary |
| 10:08 AM | Dr. Alexander Makarov | Thermo Fisher Scientific, Germany | Plenary |
| 10:36 AM | Dr. John Wilson | ProtiFi, USA | Keynote |
| 10:56 AM | Shourjo Ghose | Bruker | Keynote |
| TEA | |||
| 11:30 AM | Prof. Arup Acharjee | University of Allahabad | ECR |
| 11:40 AM | Prof. Jochen Schwenk | Scilife Lab, Sweden | Keynote (Online) |
| 12:00 Noon | Prof. Hanno Steen | Harvard Medical School, USA | Plenary |
| 12:28 PM | Dr. Prashant Kumar | Karkinos Healthcare Pvt. Ltd | Plenary |
| LUNCH | |||
| 2:00 PM | Prof. Mukunda Goswami | Central Institute of Fisheries Education | ECR |
| 2:10 PM | Prof. Sumit K. Singh | Indian Institute of Technology (BHU) Varanasi | ECR |
| 2:20 PM | Prof. Kartiki Desai | National Institute of Biomedical Genomics | ECR |
| 2:30 PM | Prof. Graham Ball | Anglia Ruskin University, UK | Plenary |
| 2:58 PM | Mr. Brendan MacLean | University of Washington, USA | Plenary |
| 3:28 PM | Prof. Robert Moritz | Institute for Systems Biology, USA | Plenary |
| 4:10 PM | Prof. Ashutosh Kumar | Indian Institute of Technology Bombay | Invited |
| 4:25 PM | Dr. Ravi Shankara | Sun Pharmaceutical Industries Ltd. | Invited |
| 4:40 PM | Dr. Bhupesh Dewan | Zuventus Healthcare Ltd. | Invited |
| 4:55 PM | Prof. Ratnesh Jain | Institute of Chemical Technology, Mumbai | Invited |
| 5:10 PM | Dr. Harsharan Singh Bhatia | Helmholtz Munich, Germany | Invited |
| 5:25 PM | Dr. Jubilee Purkayastha | Defence Research and Development Organisation (DRDO), Delhi | Plenary (Online) |
| 5:35 PM | Prof. Sandipan Ray | Indian Institute of Technology Hyderabad | ECR |
| 5:45 PM | Dr. Ankit Jain | National Centre for Biological Sciences (NCBS) | ECR |
| 5:55 PM | Student presentations (3 X 5) | Student | |
| 6:10 PM | Prof. Sanjeeva Srivastava | Indian Institute of Technology Bombay | Closing Ceremony |
SPONSORS

Exhibits

Exhibits on first floor are now filling fast…
STUDENT’S CORNER
- Oral and Poster presentations at conference (19th and 20th Feb, 2023)
- Poster presentations during workshop (17th and 18th Feb, 2023)
- Accommodation in IIT Bombay hostels (Limited availability, first come, first serve)
- Meet academia and industry experts. Networking and job opportunities.
- 1 on 1 mentorship from the keynote speakers
- Get a chance to understand the process of scientific publication from editors of reputed journals.
- Visit the Department of Biosciences and Bioengineering, Proteomics Lab and Mass Facility, IIT Bombay. Opportunities for informal interaction if you are staying on campus.
- In addition to the intense scientific sessions, there are also additional opportunities for social and cultural activities.