Regulation of Fate of Cellular Substrates
Protein-Protein interactions have been implicated in being responsible for major cellular activities ranging from cellular transduction to cellular degradation.Our lab has been focussed on understanding the cellular mechanisms associated with proteasomal degradation, especially, E3 ligases assemblies. Comprising of an intricate protein-protein network, these assemblies are capable of carrying out highly selective interactions culpable for maintaining cellular homeostasis.
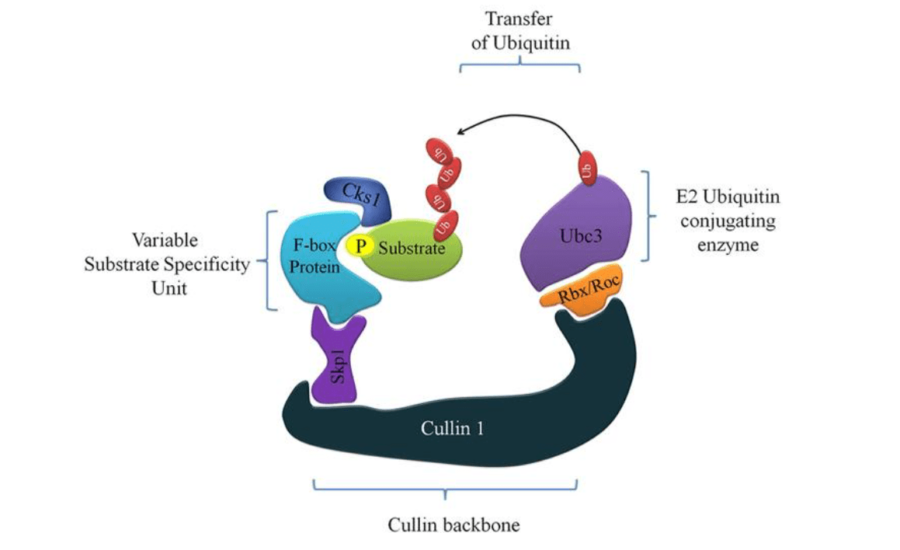
Skp1-Cullin1-Fbox (SCF) E3 ligase is a multi-protein enzyme complex responsible for the final transfer of ubiquitin to the target substrate. The diverse substrates ubiquitinated by the SCF complexes are implicated in cell cycle, DNA damage, cancer, along with several other diseases. S-phase kinase-associated protein 1 (Skp1) is the adapter protein in SCF complex which recognizes and binds to the F-box protein (FBP). FBP sequentially binds and presents the substrate fated for degradation to the rest of the E3 ligase. There are 69 different FBP’s in our body which determine the substrate specificity of SCF complexes. Skp1 recognises all these 69 F-box proteins via a single interface.
There are 69 different FBP’s in our body which determine the substrate specificity of SCF complexes. Skp1 recognises all these 69 F-box proteins via a single interface.
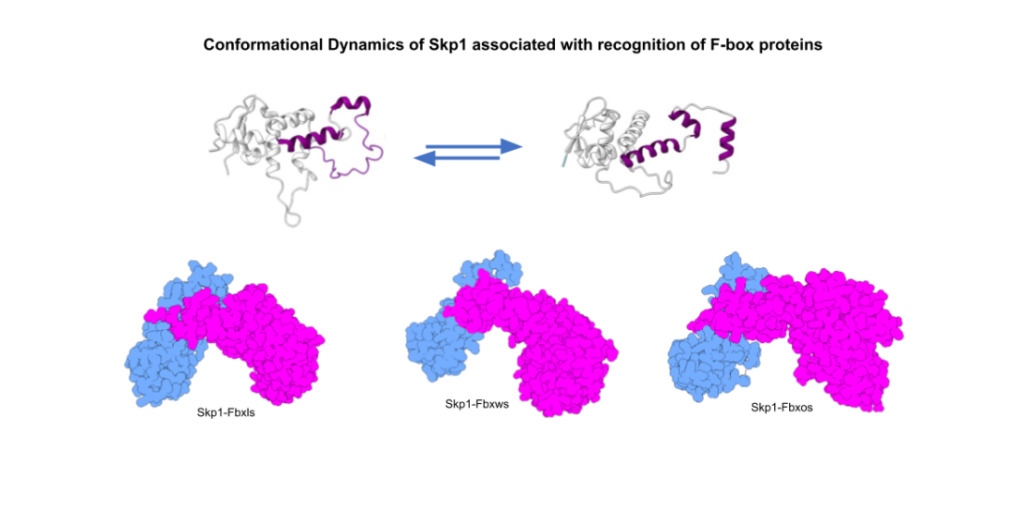
Skp1 promiscuity is the crux of our questions and raises questions on the dynamic nature of conformational transition and molecular recognition exhibited by Skp1.Role of the identified Intrinsically Disordered Region in Skp1 and its role in subsequent interaction is underway utilising Nuclear Magnetic Resonance (NMR) and Molecular Dynamics (MD)
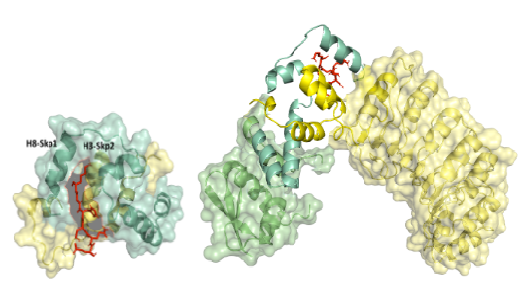
Owing to their role in several diseases, the Skp1-Fbox domain interface represents a specific drug target. Skp1 has previously been co-crystallized with different F-box proteins; however interaction and recognition dynamics of the Skp1-Fbox interface is not understood yet. We are currently investigating structure and the unique dynamics of Skp1-Fbox interface. Our goal is to develop interface selective inhibitors for Skp1-Fbox protein-protein interface. In most cases, protein-protein interaction (PPI) interfaces, are somewhat difficult to be inhibited as is indicated by several major small-molecule based designs and therefore, require alternate strategies for inhibition. We aim to design Peptide-based inhibitors, that would be better suited to fit the unconventional binding pockets of the PPI and are suitably poised to have less off-targets effects.
Publications:
1. Bhattacharya, Amrita and Shukla, Vaibhav, Preeti, Kumar,Praveen and Kumar, Ashutosh, Disorder in Human Skp1 Structure is the Key to its Adaptability to Bind Many Different Proteins in the Scf Complex Assembly. Available at SSRN: https://ssrn.com/abstract=4058896 or http://dx.doi.org/10.2139/ssrn.4058896
2. Chandra Dantu, S.; Nathubhai Kachariya, N.; Kumar, A Molecular Dynamics Simulations Elucidate the Mode of Protein Recognition by Skp1 and the F‐box Domain in the SCF Complex Proteins 2016,84(1):159-71. doi:10.1002/prot.24963
3. Kachariya, N. N.; Dantu, S. C.; Kumar, A Backbone and Side-Chain Assignments of Human Cell Cycle Regulatory Protein S-Phase Kinase-Associated Protein-1 Biomol. NMR Assign.2016,10(2):351-5. doi:10.1007/s12104-016-9699-2
Lab Members working on the various aspects of Regulation of Fates of Cellular Substrates include: Dr. Anomitra Dey,Simran Tolani, Preeti and Debarghya Mitra